The genome of a landlocked Atlantic salmon Salmo salar characterized through high-throughput sequencing


Опубликована Окт. 31, 2016
Последнее обновление статьи Авг. 21, 2022
Abstract
A wide range of life-history tactics can be found within salmonid fish. The genetic basis for these adaptations remains largely unknown, but we have sought to investigate any large scale genetic changes associated with a non-anadromous life cycle. After the most recent ice age (approximately 9,500 years ago), some populations of Atlantic salmon Salmo salar L., were trapped in fresh water and developed into isolated landlocked populations that managed to complete a full life cycle without ever reaching the marine environment. To explore whether this transition was accompanied by gene-loss events, high-throughput sequencing of a non-migratory Namsblank (‘småblank’), an Atlantic salmon from the river Namsen in Norway, was performed. There were no indications of loss of coding regions and a phylogenetic analysis based on the mitochondrial genome revealed a close genetic relationship between anadromous Atlantic salmon and Namsblank. Lack of large-scale genomic changes suggests that fine-scale (epi)genetic alterations and population genetics processes underlie adaptation to the landlocked life-style.
Ключевые слова
Illumina., salmon, gene-loss, Landlocked
INTRODUCTION
Salmonidae exhibit a tremendous diversity of phenotypic traits both at the interspecific and intraspecific levels. Variation in its phenotypic traits generally relates to reproductive strategies, mobility associated with foraging tactics, habitat selection within lakes, and tendency for anadromy. Life-history variation in salmonids appears to be influenced by complex interactions involving the endocrine system, environmental parameters (temperature, photoperiod, feeding resources), individual characteristics (sex, age, or size), and their genetic background (Hendry et al., 2004; McCormick, 2009). The genetic determinants of these life-history tactics are still largely uncovered.
Most Atlantic salmon Salmo salar L. have an anadromous life cycle in which they hatch in fresh water, migrate to sea as juveniles, and return to fresh water to spawn and die. Migration towards the sea is triggered by light-induced hormonal burst in their first or second spring (Björnsson et al., 2011). Prior to, and during, this journey, they undergo smoltification. This metamorphosis represents a set of very diverse morphological, behavioral, metabolic, endocrine and physiological shifts. Many of these developmental transitions are necessary for successful acclimation to the seawater environment, including changes in osmotic regulation and induced growth rate (Björnsson et al., 2011; Dufour et al., 2012). The timing of this transition is triggered by internal information (metabolic status, age, etc.) and by environmental cues such as light and temperature. Some of these adaptations include heritable traits subject to natural selection (Hara et al., 2007; Nichols et al., 2008; Duston et al., 2011).
Entire populations of Atlantic salmon also complete their life cycle in fresh water. Such populations are found in lakes and rivers in several countries of the northern hemisphere, including USA, Canada, Finland, Sweden, Russia, and Norway (Kazakov, 1992; Garcia de Leaniz et al., 2007; King et al., 2007). Most populations represent “glaciomarine relicts” trapped by rising land and dropping sea levels following the last ice age some 10,000 years ago (Verspoor et al., 2007). Following these isolation events, non-migratory salmon populations adapted differently, depending on the environmental conditions in their freshwater habitat and developed new phenotypes. Although lakes may house larger specimens of Atlantic salmon (> 10 kg), the salmon trapped in rivers have stayed small probably due to limited space and stronger competition for food (Berg, 1985; Gibson et al., 1996). In some aquatic taxa, the loss of a marine phase has even been linked to speciation events (Waters and Wallis, 2001).
The landlocked Atlantic salmon of the Norwegian river Namsen, Namsblank, constitutes an island population that has been isolated for approximately 9,500 years (Frankham, 1997). The habitat area available for the Namsblank population includes a 75 km stretch of the upper part of the river Namsen that together with the lower reaches of a number of tributaries amounts to 12.5 km2 of water-covered area (Sandlund et al., 2014). Effective population size estimates indicate a few thousand Namsblank fish in total, and Namsblank represents a unique cluster among Atlantic salmon populations with regard to biological and genetic characteristics. Genetic analyses suggest that Namsblank can be further divided into different subpopulations with estimated effective population sizes of a few hundred fish (Sandlund et al., 2014).
Namsblank individuals are among the smallest relict Atlantic salmon in the world and rarely grow larger than 30 cm (300 g). Fish keep the visual appearance of a young salmon parr (Berg, 1956; 1985), which may indicate that a range of developmental changes normally occurring early in life of anadromous (migrating) Atlantic salmon are not induced, along with the need to migrate, smoltify and grow to full size (Berg, 1985). The loss of such features can be linked to genetic, epigenetic or environmental differences (Garcia de Leaniz et al., 2007).
As a result of a whole-genome duplication (WGD) event 25 to 100 million years ago, salmonids have a tetraploid genome that is currently in the process of rediploidization (Allendorf et al., 1984; Macqueen and Johnston, 2014). It has been suggested that polyploids possess greater evolutionary potential than their diploid progenitors, as genetic redundancy creates opportunities for duplicated genes to be lost or to diverge and acquire new functions (Lien et al., 2016), in addition to epigenetic remodeling affecting gene expression (Comai, 2005). Thus, polyploidy may allow organisms to evolve faster or in novel directions compared to their diploid progenitors, leading to increased evolutionary success due to improved ability to diversify and adapt to new niches.
In this work, we wanted to investigate whether the Namsblank phenotype is linked to genetic changes when compared with anadromous Atlantic salmon. Given the evolutionary history of Namsblank and the nature of the salmonid genome, it is not unreasonable to expect both large-scale and small-scale genetic changes, either through genetic drift or as a result of changes in selection pressure. As an initial screen for large-scale genetic changes, we used high-throughput sequencing as an approach to identify gene-loss events in the Namsblank genome. In order to estimate the evolutionary distance between migratory Atlantic salmon and the landlocked Namsblank, a phylogenetic analysis was also performed based on the complete mitochondrial genome. This represents the first sequence data reported from a landlocked Atlantic salmon population.
MATERIALS AND METHODS
DNA was isolated from kidney tissue of an adult, male Namsblank specimen (Figure 1) using the DNeasy Blood & Tissue Kit (QIAGEN AB, Oslo, Norway) after homogenization using a glass mortar and pestle. The extracted DNA was fragmented and a 3 kb mate-pair library prepared. Sequencing (two lanes) was performed using the Illumina HiSeq 2000 platform (GATC Biotech AB, Konstanz, Germany).
Complete Atlantic salmon mitochondrial genome sequences were downloaded from GenBank (National Center for Biotechnology Information) and aligned using the Clustal W algorithm (Thompson et al., 1994). Sequence reads matching the mitochondrial genome database were identified using the Mega BLAST algorithm with default search parameters (word size 28) (Zhang et al., 2000). Matching reads were extracted from the Illumina read database and aligned with a complete reference Atlantic salmon mitochondrial genome that showed a high degree of sequence similarity with our data (GenBank accession number JQ390056) using the CLC Genomics Workbench, version 6.0.4 (CLC bio, Aarhus, Denmark), and a consensus sequence was generated.
For a comparative gene-content analysis, all Atlantic salmon mRNA sequences deposited in GenBank were downloaded (48,205). Mega BLAST was used (with the same settings as above) to align the Illumina sequence reads obtained with the downloaded salmon mRNA data. Transcript sequences for which we had matching reads were successively removed in order to identify regions of possible gene-loss. Primers were designed for a set of gene-loss candidates using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/). PCR was performed according to the manufacturer’s instructions using Tfi DNA polymerase (Life Technologies, Oslo, Norway). The PCR cycling conditions were 94°C for 2 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min, and a final extension step at 72°C for 10 min. Amplified products were analyzed using an Agilent 2100 Bioanalyzer using a DNA 1000 kit (Agilent Technologies). PCRs were performed using DNA from the same Namsblank specimen as was originally submitted for sequencing in addition to a small panel of anadromous Atlantic salmon specimens samples from Norwegian (wild and farmed) and Irish (farmed) locations.
RESULTS
The Illumina sequencing yielded a total of 381,479,528 reads (length: 101 bases, the two sets of paired-end reads were submitted to the European Nucleotide Archive as study PRJEB15379). The haploid genome of Atlantic salmon has been reported to be approximately 2.97 gigabases (Lien et al., 2016) and the theoretical coverage of the Atlantic salmon genome should thus be approximately 12X.
When comparing our Illumina data set to our database of salmon mRNA sequences, 47,924 (99.4%) of the transcript entries had a match with one or more of our Illumina reads, leaving 281 mRNA sequences for which no matching reads were found. This number was reduced to 250 sequences by excluding identical entries (Supplementary Tables S1 and S2). Sequences were ranked according to length and primers were designed for the thirteen longest accessions having journal references (Supplementary Table S2). In order to ensure that primers were located in exonic regions, candidate mRNA sequences were mapped against available genomic sequences from Atlantic salmon or other fish species. In addition, a positive control PCR was designed using the 18S rRNA gene as target locus. PCR was performed using genomic DNA. All PCRs were negative (except the positive control).
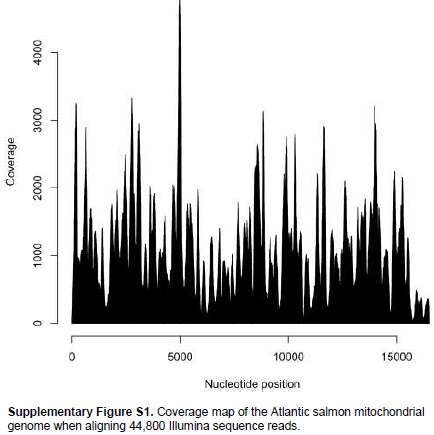
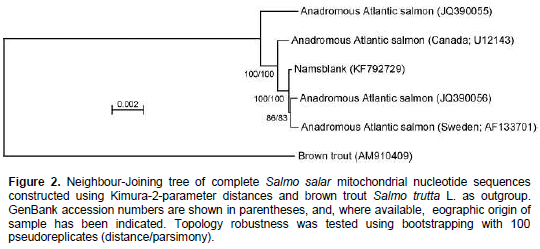
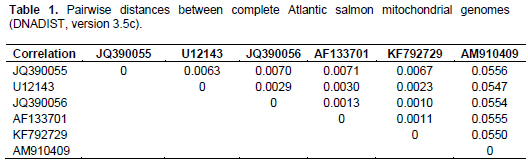
When aligning 44,800 mitochondrial reads with a reference Atlantic salmon mitochondrial genome, a significant bias in coverage was observed (Supplementary Figure S1). This appears to have been caused by partial degradation of the template DNA (Dr. Volker Brenner, GATC Biotech AG, personal communication). A Neighbour-Joining analysis using Kimura-2-parameter distances (SeaView, version 4.2.11; Gouy et al., 2010) of the the complete Namsblank mitochondrial genome (GenBank accession number KF792729) indicated that the genetic distance between Namsblank and anadromous strains of Atlantic salmon is small (Figure 2 and Table 1) and no mutations unique for the landlocked mitochondrial DNA sequence were observed.
DISCUSSION
Assessment of single nucleotide polymorphisms (SNPs) in mitochondrial and nuclear DNA of different populations of Atlantic salmon, including landlocked isolates, indicates a polyphyletic origin of these different subpopulations (Bourret et al., 2013). Still, all populations of Atlantic salmon are referred to as the same species. The mitochondrial genome has not gone through the same changes in ploidy as the nuclear salmonid genome, leading to a lower level of gene-content redundancy. Combined with the clonal mode of mitochondrial replication and the relatively short time of population separation, this might explain the close genetic relationship between Namsblank and anadromous Atlantic salmon populations when comparing mitochondrial genome sequences. Our dataset did not allow for a detailed comparison between the anadromous Atlantic salmon genome and our Namsblank specimen. Our strategy of discarding sequences based on a cut-off rule that requires 100% match only in a 28 basepair region may also have led to the indiscriminate filtering of paralogous genes and not just homologous alleles, so even loss of all paralogs of a given gene could be missed. When mining for salmon sequence data in the NCBI database, we found numerous examples of obvious errors and apparent contamination, for instance NM_001140589 (‘Salmo salar Photosystem II 10 kDa polypeptide, chloroplast (psbr)), mRNA’ and BT046286 (‘Salmo salar clone ssal-evf-563-177 Photosystem II 10 kDa polypeptide, chloroplast precursor putative mRNA, complete cds’). Both of these examples are likely the result of plant material contamination of the sequenced samples, as subsequent nucleotide similarity searches revealed that both entries appeared to in fact stem from plant DNA. Our hypothesis for the lack of PCR products is thus due to either erroneously annotated GenBank accessions or mRNA sequences derived from poor quality data in our salmon transcript database. It is also possible that binding sites for some of the primers that were based on gene structure data from other fish species did not reside within the intended exonic regions. The lack of positive control material also may have introduced some false negatives due to poor primer design, but we consider this unlikely as the primers were designed using standardized software and PCRs were run using robust conditions.
A situation analogous to the isolation of Atlantic salmon populations in freshwater localities could be the Mexican tetra Astyanax mexicanus (Filippi). This species has given rise to several blind cave subpopulations that have lived in complete darkness and isolation for at least 10,000 years. In these populations, several loss-of-function mutations have been described (Jeffery, 2001) and mutations in genes such as Oca2 and Mc1r have been linked directly to an observed reduction in pigmentation (Protas et al., 2006; Gross et al., 2009). Interestingly, there even seem to have been several constructive (gain-of-function) changes, including increased number of taste buds, extra teeth and a more robust jaw structure (Jeffery, 2001). We do not exclude the possibility that there are functionally relevant genetic differences between Namsblank and anadromous Atlantic salmon, but the transition from an anadromous life cycle to isolation in freshwater may have been accompanied by more subtle, regulatory mutations or even epigenetic changes, possibly in a combinatorial fashion where multiple genes have been affected. Experimental attempts to induce smoltification in another landlocked Norwegian population (salmon from Lake Byglandsfjord, ‘Byglandsbleke‘) revealed reduced hormonal responses to natural spring light (plasma growth hormone and cortisol) compared to anadromous salmon. The corresponding growth hormone receptor response in gills, which is known to precede gill adaptation to seawater handling, was also reduced (Nilsen et al., 2008). In a comparison between Atlantic salmon and the less seawater-adapted Arctic char, quantitative trait loci (QTLs) linked to seawater tolerance were identified in genes encoding osmoregulatory proteins, hormonal signals of smoltification and an epithelial junction protein (Norman et al., 2012). Morphological, physiological and behavioural differences that may be linked to epigenetic adaptation have been described in teleost species such as Arctic char Salvelinus alpinus L. (Adams et al., 2003; Arbour et al., 2011). Such epigenetically based phenotypic plasticity may play a role in adaptation or speciation (Pfennig et al., 2010) and could also contribute to the observed differences between landlocked and anadromous Atlantic salmon. Changes in gene-expression levels have been linked with life-history variation in salmonid fish in previous studies and a gene-expression signature for future migratory behavior has been suggested for brown trout Salmo trutta L. (Giger et al., 2008). Another interesting finding from this study is that variance in life-history seems to be a much more important factor than genetic diversity when explaining differences in gene-expression profiles. For immune-related genes, selection sweeps have been identified using SNP data in Atlantic salmon (Kjærner-Semb et al., 2016) and it is not inconceivable that similar data might indicate that similar adaptational processes have taken place in the transition from a migratory to a residential life-history.
Until a fully annotated reference genome as well as information on genetic (expression) variability on a population level is available for a large number of Atlantic salmon isolates, it will be very challenging to pinpoint the exact genetic differences that enable subpopulations to spend their entire life cycle in freshwater. However, a more detailed understanding of the underlying mechanisms should give insight into Atlantic salmon adaptive processes in general and could prove very useful when optimizing the smoltification process for Atlantic salmon reared in aquaculture, as well as for any future attempts to keep adult salmon in land-based freshwater facilities.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
ACKNOWLEDGEMENTS
The authors thank Dr. Ole Kristian Berg (Norwegian University of Science and Technology) for kindly providing the Namsblank specimen. This work was supported by internal funding from the Norwegian Veterinary Institute (Mucopath project).
REFERENCES
Adams CE, Woltering C, Alexander G (2003). Epigenetic regulation of trophic morphology through feeding behaviour in Arctic charr, Salvelinus alpinus. Biol. J. Linn. Soc. 78(1):43-49. | |
Allendorf FW, Stahl G, Ryman N (1984). Silencing of duplicate genes: a null allele polymorphism for lactate dehydrogenase in brown trout (Salmo trutta). Mol. Biol. Evol. 1(3):238-248. | |
Arbour JH, Hardie DC, Hutchings JA (2011). Morphometric and genetic analyses of two sympatric morphs of Arctic char (Salvelinus alpinus) in the Canadian High Arctic. Can. J. Zool. 89(1):19-30. | |
Berg M (1956). A relict salmon, Salmo salar L., called "småblank" from the river Namsen, North Trøndelag. Acta Borealia A. Scientia 6, 17s. | |
Berg OK (1985). The formation of non-anadromous populations of Atlantic salmon, Salmo salar L., in Europe. J. Fish Biol. 27(6):805-815. | |
Björnsson BT, Stefansson SO, McCormick SD (2011). Environmental endocrinology of salmon smoltification. Gen. Comp. Endocrinol. 170(2):290-298. | |
Bourret V, Kent MP, Primmer CR, Vasemagi A, Karlsson S, Hindar K, McGinnity P, Verspoor E, Bernatchez L, Lien, S (2013). SNP-array reveals genome-wide patterns of geographical and potential adaptive divergence across the natural range of Atlantic salmon (Salmo salar). Mol. Ecol. 22(3):532-551. | |
Comai L (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6(11):836-846. | |
Dufour S, Rousseau K, Kapoor BG (2012). Metamorphosis in fish. Enfield, NH. Boca Raton, FL, Science Publishers, distributed by CRC Press. | |
Duston J, Astatkie T, Zhang C (2011). Hypo-osmoregulatory capacity during smolting of endangered inner Bay of Fundy Atlantic salmon and other eastern Canadian stocks. Aquac. 319(1-2):221-225. | |
Frankham R (1997). Do island populations have less genetic variation than mainland populations? Heredity 78(3):311-327. | |
Garcia de Leaniz C, Fleming IA, Einum S, Verspoor E, Jordan WC, Consuegra S, Aubin-Horth N, Lajus D, Letcher BH, Youngson AF, Webb JH, Vollestad LA, Villanueva B, Ferguson A, Quinn TP (2007). A critical review of adaptive genetic variation in Atlantic salmon: implications for conservation. Biol. Rev. 82(2):173-211. | |
Gibson RJ, Williams DD, McGowan C, Davidson WS (1996). The ecology of dwarf fluvial Atlantic salmon, Salmo salar L., cohabiting with brook trout, Salvelinus fontinalis (Mitchill), in southeastern Newfoundland, Canada. Proceedings of the 5th international symposium on the ecology of fluvial fishes, Pol. Arch. Hydrobiol. 43:145-166. | |
Giger T, Excoffier L, Amstutz U, Day PJ, Champigneulle A, Hansen MM, Kelso J, Largiadèr CR (2008). Population transcriptomics of life-history variation in the genus Salmo. Mol. Ecol. 17(13):3095-108. | |
Gouy M, Guindon S, Gascuel O (2010). SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27(2):221-224. | |
Gross JB, Borowsky R, Tabin, CJ (2009). A Novel Role for Mc1r in the Parallel Evolution of Depigmentation in Independent Populations of the Cavefish Astyanax mexicanus. PLoS Genet. 5(1):e1000326. | |
Hara T, Nagase T, Ozaki A, Sakamoto T, Okamoto N, Danzmann RG, Tokuhara T, Kuwada T (2007). A genetic linkage map of amago salmon (Oncorhynchus masou ishikawae) and mapping of quantitative trait loci associated with smoltification. Aquac. 272(1):S266-S267. | |
Hardie DC, Hebert PD (2003). The nucleotypic effects of cellular DNA content in cartilaginous and ray-finned fishes. Genome 46(4):683-706. | |
Hendry AP, Morbey YE, Berg OK, Wenburg JK (2004). Adaptive variation in senescence: reproductive lifespan in a wild salmon population. Proc. R. Soc. Lond. B Biol. Sci. 271(1536):259-66. | |
Jeffery WR (2001). Cavefish as a model system in evolutionary developmental biology. Dev. Biol. 231(1):1-12. | |
Kazakov RV (1992). Distribution of Atlantic salmon Salmo salar L., in freshwater bodies of Europe. Aquac. Fish. Manag. 23(4):461-475. | |
King TL, Verspoor E, Spidle AP, Gross R, Phillips RB, Koljonen ML, Sanchez JA, Morrison C (2007). Biodiversity and population structure; The Atlantic Salmon: Genetics, Conservation and Management. Blackwell, Oxford, pp. 117-166. | |
Kjærner-Semb E, Ayllon F, Furmanek T, Wennevik V, Dahle G, Niemelä E, Ozerov M, Vähä JP, Glover KA, Rubin CJ, Wargelius A, Edvardsen RB (2016). Atlantic salmon populations reveal adaptive divergence of immune related genes - a duplicated genome under selection. BMC Genomics 17(1):610. | |
Lien S, Koop BF, Sandve SR, Miller JR, Kent MP, Nome T, Hvidsten TR, Leong JS, Minkley DR, Zimin A, Grammes F, Grove H, Gjuvsland A, Walenz B, Hermansen RA, von Schalburg K, Rondeau EB, Di Genova A, Samy JK, Olav Vik J, Vigeland MD, Caler L, Grimholt U, Jentoft S, Våge DI, de Jong P, Moen T, Baranski M, Palti Y, Smith DR, Yorke JA, Nederbragt AJ, Tooming-Klunderud A, Jakobsen KS, Jiang X, Fan D, Hu Y, Liberles DA, Vidal R, Iturra P, Jones SJ, Jonassen I, Maass A, Omholt SW, Davidson WS (2016). The Atlantic salmon genome provides insights into rediploidization. Nat. 533(7602):200-205. | |
Macqueen DJ, Johnston IA (2014). A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc. R. Soc. Lond. B Biol. Sci. 281(1778):20132881. | |
McCormick SD (2009). Evolution of the hormonal control of animal performance: Insights from the seaward migration of salmon. Integr. Comp. Biol. 49(4):408-22. | |
Nichols KM, Edo AF, Wheeler PA, Thorgaard GH (2008). The genetic basis of smoltification-related traits in Oncorhynchus mykiss. Genet. 179(3):1559-1575. | |
Nilsen TO, Ebbesson LO, Kiilerich P, Bjornsson BT, Madsen, SS, McCormick SD, Stefansson SO (2008). Endocrine systems in juvenile anadromous and landlocked Atlantic salmon (Salmo salar): seasonal development and seawater acclimation. Gen.Comp. Endocrinol. 155(3):762-772. | |
Norman JD, Robinson M, Glebe B, Ferguson MM, Danzmann RG (2012). Genomic arrangement of salinity tolerance QTLs in salmonids: a comparative analysis of Atlantic salmon (Salmo salar) with Arctic charr (Salvelinus alpinus) and rainbow trout (Oncorhynchus mykiss). BMC Genomics 13:420. | |
Pfennig DW, Wund MA, Snell-Rood EC, Cruickshank T, Schlichting CD, Moczek AP (2010). Phenotypic plasticity's impacts on diversification and speciation. Trend. Ecol. Evol. 25(8):459-467. | |
Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon LI, Borowsky R, Tabin CJ (2006). Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat. Genet. 38(1):107-111. | |
Sandlund OT, Karlsson S, Thorstad EB, Berg OK, Kent MP, Norum IC, Hindar K (2014). Spatial and temporal genetic structure of a river-resident Atlantic salmon (Salmo salar) after millennia of isolation. Ecol. Evol. 4(9):1538-1554. | |
Thompson JD, Higgins DG, Gibson TJ (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673-4680. | |
Verspoor E, Stradmeyer L, Nielsen JL and the Atlantic Salmon Trust (2007). The Atlantic salmon: genetics, conservation and management. Blackwell, Oxford. | |
Waters JM, Wallis GP (2001). Cladogenesis and loss of the marine life-history phase in freshwater galaxiid fishes (Osmeriformes: Galaxiidae). Evolution 55(3):587-597. | |
Zhang Z, Schwartz S, Wagner L, Miller W (2000). A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7(1-2):203-214. | |